Summary of VEGA
DRAFT POST
This post is a rough summary of the VEGA architecture presented in VEGA is an interpretable generative model for inferring biological network activity in single-cell transcriptomics, published by Seninge et al. in Nature Communications. I’m hoping that writing it up here will allow me to understand it better. If you understand this work and its position in the wider context of variational autoencoders better than me and notice mistakes in my write-up, then please do get in touch via the link at the bottom of this page!
VEGA (“VAE Enhanced by Gene Annotations”) is a variational autoencoder (VAE) built to reduce dimensionality of a single-cell transcriptomic dataset by more meaningfully capture the biological information driving variance within it. This means that, in order to understand what VEGA is, we must first understand VAEs. The first part of this post will deal with that and then we will revisit VEGA.
Variational autoencoders (VAEs)
This section is largely based on / inspired by a nice post by Francesco Franco
An autoencoder (AE) is a pair of connected neural networks (an encoder and a decoder) that aims to find a lower-dimensionality representation of a dataset. As you might expect, the encoder encodes data points and the decoder decodes them again. Graphically:
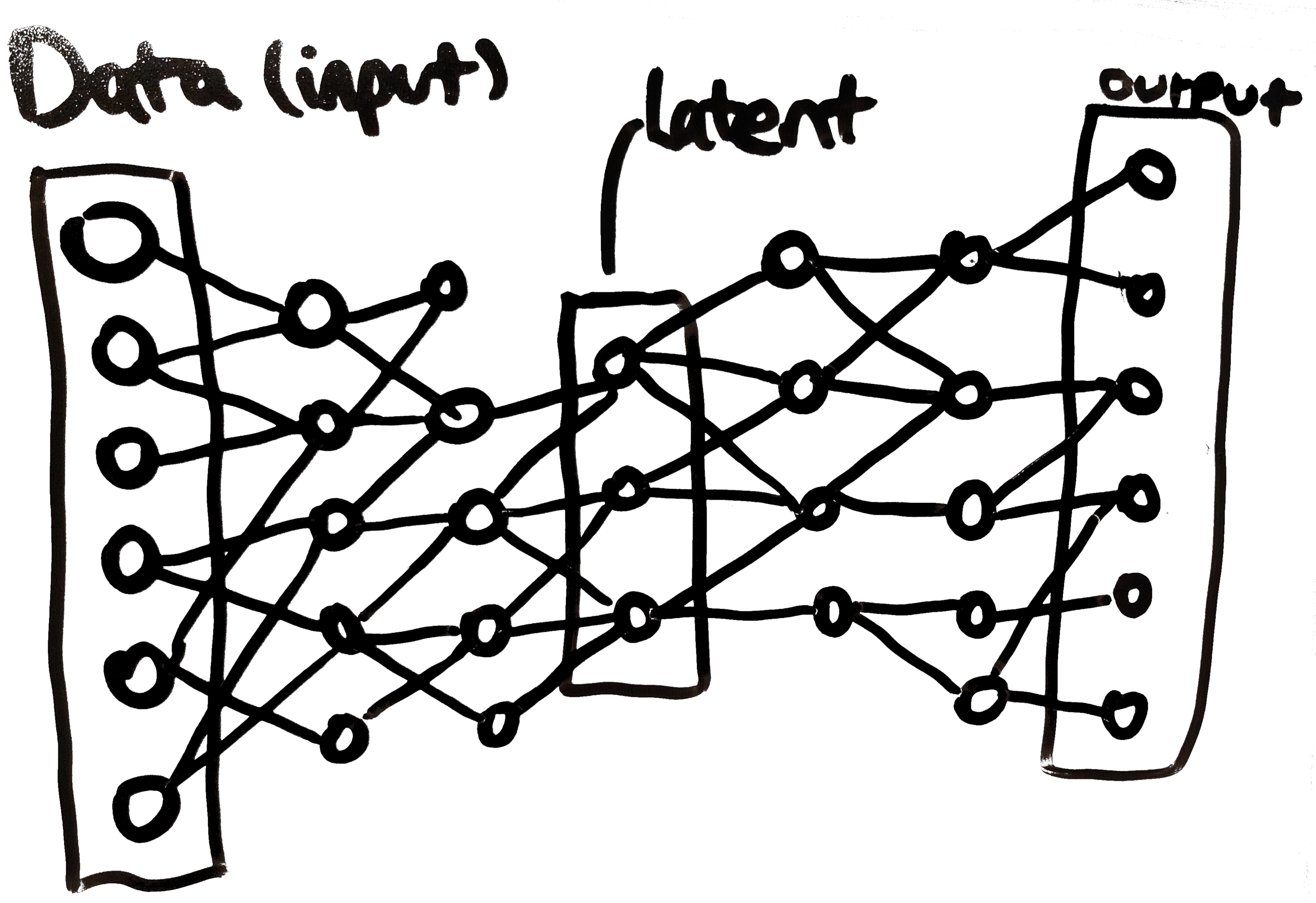
(yes, there is a node connected to nothing in the second hidden layer 🤦 - this is just a diagram bug…) The bottleneck layer (labelled “latent” above) is the reduced dimension representation of the instance used as input on the left of the diagram. The decoder portion of the model takes the latent representation and attempts to recreate the original instance. If a dataset is able to pass through the autoencoder with minimal reconstruction loss (i.e. artefacts introduced by the compression), then we know that the latent representation in the middle, when coupled with the trained decoder network, is a reasonable compressed representation of the data.
A decoder can be used to generate novel instances following the distribution of the training data by ‘decoding’ a point in the latent space (i.e. assigning arbitrary values latent layer). However, it turns out that a standard autoencoder does a bad job of generating realistic data: although the latent space surrounding training examples yield realistic novel data, the space far from training data does not. Ideally, we would develop an autoencoder that learns a latent representation that generates realistic data everywhere.
Variational autoencoders (VAEs) are able to achieve this aim by smoothing the latent space between the training points. Instead of learning single representations for each instance, a VAE instead learns a probability distribution over the latent space. The latent nodes correspond to the parameters of this distribution. Thus, in a VAE, the input to the decoder is a sample from this learned probability distribution. Using probability distributions smooths the areas around the training instances, leading to better generation.
[I need to detail this much better – e.g. why does a VAE yield smoother latent space?]
VEGA
We are now in a position to better understand VEGA. Instead of learning a latent representation without biological meaning, VEGA attempts to learn a latent representation of a single-cell transcriptomic dataset that captures the expression of gene sets by each cell. It associates each latent node of the VAE with a gene set and uses a decoder with a single, linear layer in which an output node (i.e. a gene) is only connected to the latent nodes representing gene sets to which it belongs. In this way, cells that have similar latent representations in VEGA likely express similar gene sets. The following a schematic for VEGA:
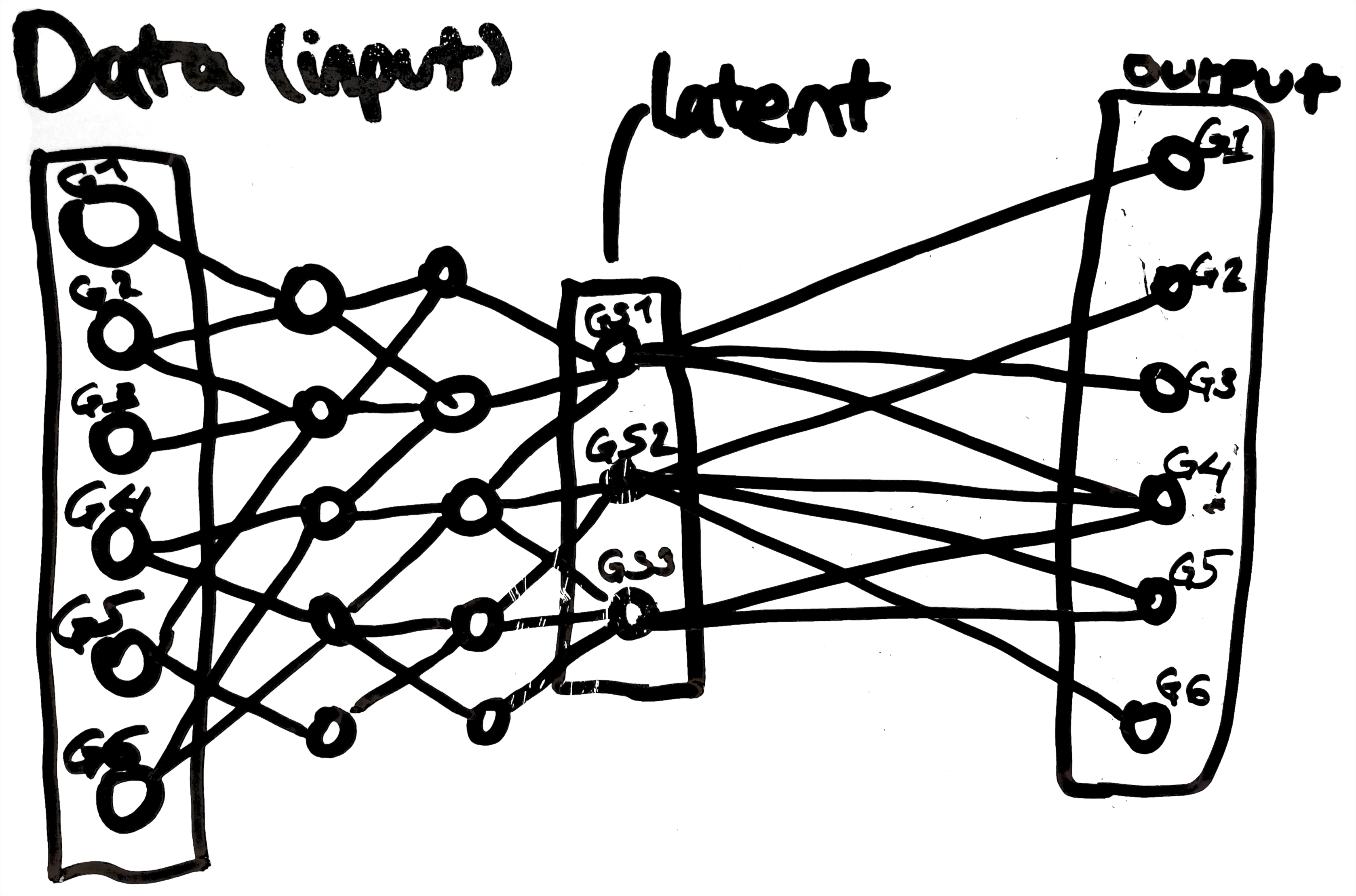
The input and output nodes correspond to genes G1-G6 and the latent nodes correspond to gene sets GS1-GS3.
Any comments?
Feel free to submit an issue here.
Disclaimer: The content in this post is provided as is, without warranty of any kind. I make no guarantees about the completeness, reliability, or accuracy of the code, tips, or advice presented. Any action you take based on this content is strictly at your own risk. I will not be liable for any losses, damages, or issues arising from the use or misuse of this information, including (but not limited to) loss of data, system failures, or security vulnerabilities. Always test code and approaches in a safe environment before deploying them in production.
Enjoy Reading This Article?
Here are some more articles you might like to read next: